Protein/peptide sequencing
Figure 1: PPSQ protein/peptide sequencer
Protein/Peptide Sequencing

Figure 1: PPSQ protein/peptide sequencer
The laboratory offers sensitive N-terminal protein/peptide sequencing using a recently installed Shimadzu PPSQ 33B sequencer (Figure 1) at IBBR-Napoli (Figure 2). The method is based on the Edman degradation reaction which labels the N-terminal amino acid residues within a polypeptide such that they can be cleaved from the chain in a stepwise manner (Figure 3). The benefits of using Edman degradation for protein sequencing are:
- Guaranteed N-terminal sequence of proteins;
- No sample pre-treatment required:
- Differentiation of isobaric amino acids (leucine and isoleucine):
- Sequencing of unknown proteins not registered in databases.
Preparing samples for sequencing

Figure 2: PPSQ 33 B at work at IBBR-CNR-Napoli
- lyophilized
- in solution
- immobilized on PVDF membrane (a Coomassie stainable amount)
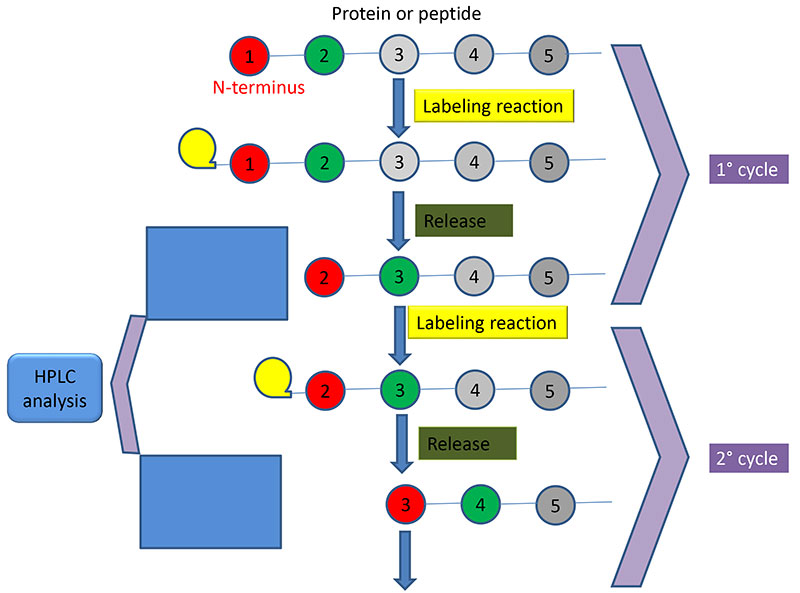
Figure 3: Schematics of the workflow used by the sequencer.




